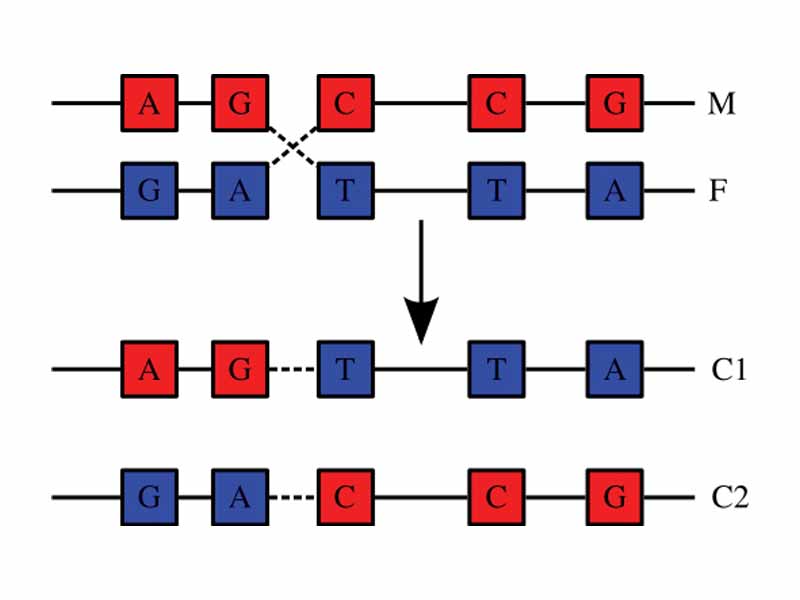
Recombination and mutation are processes leading to changes in genomes over time. The terms mutation and recombination are generally used to distinguish between small and large scale changes to the genome. Changes that happen to DNA in somatic cells are usually not heritable whereas changes that occur to the DNA in germline cells may be inherited. Recombinant DNA technologies in the lab are saved for treatment in the section on the Molecular Biology Laboratory.
WikiPremed Resources
Recombination and Mutation Images
Conceptual Vocabulary Self-Test
Basic Terms Crossword Puzzle
Basic Puzzle Solution
Conceptual Vocabulary for Recombination and Mutation
Genetic recombination is the process by which a strand of DNA is broken and then joined to the end of a different DNA molecule.
Crossing over is the process by which two chromosomes, paired up during prophase 1 of meiosis, exchange some portion of their DNA.
Homologous recombination involves the alignment of similar sequences, a crossover between the aligned DNA strands, and breaking and repair of the DNA to produce an exchange of material between the strands.
Mutations are changes to the base pair sequence of the genetic material of an organism.
A mutagen is a physical or chemical agent that changes the genetic information of an organism.
Genetic insertion is the addition of one or more nucleotide base pairs into a genetic sequence.
A deletion is a mutation in which a part of a chromosome or a sequence of DNA is missing.
An inversion is a chromosome rearrangement in which a segment of a chromosome is reversed end to end.
A frameshift mutation is a genetic mutation caused by inserts or deletes from a DNA sequence of a number of nucleotides not evenly divisible by three.
Transposons are sequences of DNA that can move around to different positions within the genome of a single cell, a process called transposition.
A germline mutation is any detectable, heritable variation in the lineage of germ cells.
DNA repair refers to a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome.
A point mutation, or single base substitution, is a type of mutation that causes the replacement of a single base nucleotide with another nucleotide.
A nonsense mutation is a point mutation in a sequence of DNA that results in a premature stop codon, or a nonsense codon in the transcribed mRNA, and possibly a truncated, and often nonfunctional protein product.
Missense mutations or nonsynonymous mutations are types of point mutations where a single nucleotide is changed to cause substitution of a different amino acid.
Silent mutations are DNA mutations that do not result in a change to the amino acid sequence of a protein.
Bacterial conjugation is the transfer of genetic material between bacteria through direct cell-to-cell contact.
Transduction is the process by which bacterial DNA is moved from one bacterium to another by a virus.
Gene conversion may occur during meiotic division through a process by which DNA sequence information is transferred from one DNA helix to another DNA helix, whose sequence is altered.
An insertion sequence is a short DNA sequence that acts as a simple transposable element.
Retrotransposons are genetic elements that can amplify themselves in a genome and are ubiquitous components of the DNA of many eukaryotic organisms.
A chromosome translocation is a chromosome abnormality caused by rearrangement of parts between nonhomologous chromosomes.
Transfection describes the introduction of foreign material into eukaryotic cells, which may occur with genetic material such as supercoiled plasmid DNA.
Horizontal or lateral gene transfer is any process in which an organism transfers genetic material to another cell that is not its offspring.
Mobile genetic elements are a type of DNA that can move around within the genome including transposons, plasmids, bacteriophage elements and group II introns.
Griffith's experiment, conducted in 1928, was one of the first experiments suggesting that bacteria are capable of transferring genetic information though a process known as transformation.
Gene duplication is any creation of a repeat of a region of DNA that contains a gene.
Polyploidy is the condition of some biological cells and organisms manifested by the presence of more than two homologous sets of chromosomes.
Transposase is an enzyme that binds to single-stranded DNA and can incorporate it into genomic DNA.
The mutation rate is the chance of a mutation occurring in an organism or gene in each generation.
An inverted repeat is a sequence of nucleotides that is the reversed complement of another sequence further downstream.
Long terminal repeats are found in retroviral DNA flanking functional genes. They encode a specific transposase called integrase which mediates integration of viral DNA into the host chromosome.
A centimorgan (abbreviated cM) is a unit of recombinant frequency for measuring genetic linkage.
Muller's ratchet is the name given to the process by which the genomes of an asexual population accumulate deleterious mutations in an irreversible manner.
Base excision repair is a cellular mechanism that can repair damaged DNA during DNA replication involving flipping the mutated base out of the DNA helix and repairing the base alone.
Nucleotide excision repair is a DNA repair mechanism involves the removal and replacement of short single-stranded DNA segments.
Mismatch repair is a system for recognizing and repairing the erroneous insertion, deletion and mis-incorporation of bases that can arise during DNA replication and recombination.
A transition is a mutation changing a purine to another purine nucleotide or a pyrimidine to another pyrimidine nucleotide.
A transversion is a type of point mutation refering to the substitution of a purine for a pyrimidine or vice versa.
Depurination is DNA alteration in which the hydrolysis of a purine base from the deoxyribose-phosphate backbone occurs.
A base analog is a chemical that can substitute for a normal nucleobase in nucleic acids.
Intercalation is the reversible inclusion of a molecule (or group) between two other molecules (or groups).
A fusion gene is a hybrid gene formed from two previously separate genes which can occur as the result of a translocation, interstitial deletion, or chromosomal inversion.
The mobilome is the total of all mobile genetic elements in a genome.
Replicative transposition is a mechanism of transposition in which the transposable element is duplicated during the reaction, so that the transposing entity is a copy of the original element.
An AP site, meaning apurinic and apyrimidinic, is a location in DNA that does not have either a purine or pyrimidine base, usually due to DNA damage.
Non-homologous end joining is a pathway that can be used to repair double-strand breaks in DNA.
Loss of heterozygosity in a cell represents the loss of one parent's contribution to part of the cell's genome.
An AP endonuclease is an enzyme that cuts a strand of DNA on the five prime side of an AP site, as part of DNA base excision repair.
UvrABC endonuclease is a multienzyme complex in E.coli bacteria involved in DNA repair mechanism by nucleotide excision repair and it is therefore sometimes called and excinuclease.
Triparental mating is a form of Bacterial conjugation where a conjugative plasmid present in one bacterial strain assists the transfer of a mobilizable plasmid present in a second bacterial strain into a third bacterial strain.
 The WikiPremed MCAT Course is a comprehensive course in the undergraduate level general sciences. Undergraduate level physics, chemistry, organic chemistry and biology are presented by this course as a unified whole within a spiraling curriculum. Please read our policies on Privacy and Shipping & Returns. Contact Us. MCAT is a registered trademark of the Association of American Medical Colleges, which does not endorse the WikiPremed Course. WikiPremed offers the customers of our publications or our teaching services no guarantees regarding eventual performance on the MCAT. The WikiPremed MCAT Course is a comprehensive course in the undergraduate level general sciences. Undergraduate level physics, chemistry, organic chemistry and biology are presented by this course as a unified whole within a spiraling curriculum. Please read our policies on Privacy and Shipping & Returns. Contact Us. MCAT is a registered trademark of the Association of American Medical Colleges, which does not endorse the WikiPremed Course. WikiPremed offers the customers of our publications or our teaching services no guarantees regarding eventual performance on the MCAT.

WikiPremed is a trademark of Wisebridge Learning Systems LLC. The work of WikiPremed is published under a Creative Commons Attribution NonCommercial ShareAlike License. There are elements of work here, such as a subset of the images in the archive from WikiPedia, that originated as GNU General Public License works, so take care to follow the unique stipulations of that license in printed reproductions. |