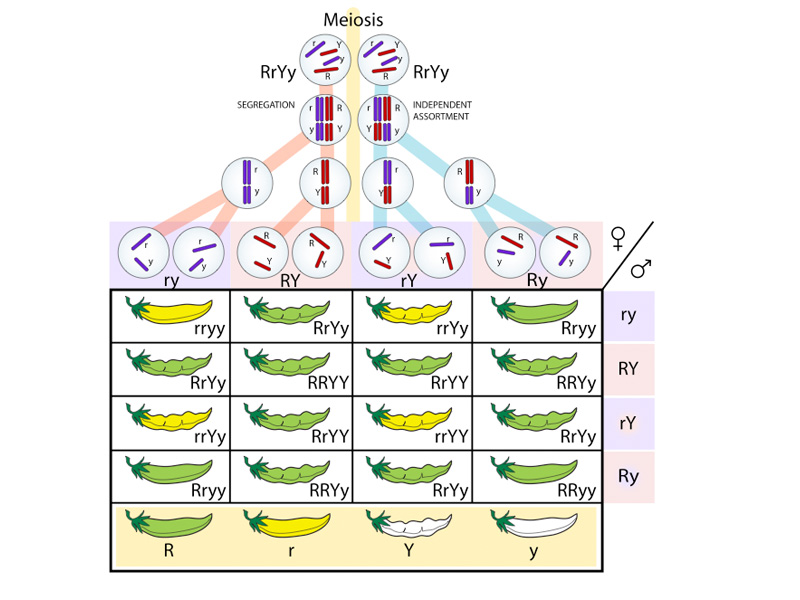
Mendelian genetics provides the basis for understanding the transmission of traits from parental organisms to their offspring. Though modified by our current understanding of the chromosomal theory of inheritance and molecular biology, Mendel's laws form the theoretical basis of our understanding of the genetics of inheritance. The law of segregation holds that during gamete formation, the alleles for each gene segregate from each other so that each gamete carries only one allele for each gene. The law of independent assortment holds that genes for different traits can segregate independently during the formation of gametes. The law of dominance holds that some alleles are dominant while others are recessive; an organism with at least one dominant allele will display the effect of the dominant allele.
WikiPremed Resources
Mendelian Genetics Images
Conceptual Vocabulary Self-Test
Basic Terms Crossword Puzzle
Basic Puzzle Solution
Conceptual Vocabulary for Transmission Genetics
An allele is a viable DNA coding that occupies a given locus on a chromosome.
The genotype describes the genetic constitution of an individual, that is the specific allelic makeup of an individual, usually with reference to a specific character under consideration.
The phenotype describes the ourward physical appearance or manifestation of a trait in an organism, as opposed to its genotype.
Gregor Mendel (1822 - 1884) was an Austrian Augustinian priest and scientist often called the father of modern genetics for his study of the inheritance of traits in pea plants.
A locus is a fixed position on a chromosome.
A dominance relationship refers to how the alleles for a locus interact to produce a phenotype.
The Punnett square is a diagram used by biologists to determine the probability of an offspring having a particular genotype.
A dihybrid cross is a cross between two individuals identically heterozygous at two loci.
A monohybrid cross is a cross between individuals who are identically heterozygous at one locus.
A pedigree chart is a chart which shows the known phenotypes for an organism and its ancestors.
A test cross crosses a homozygous recessive with an individual with an unknown genotype.
A trait is a distinct phenotypic character of an organism that may be inherited, environmentally determined or somewhere in between.
The wild type is the typical form of an organism, strain, gene, or characteristic as it occurs in nature.
F1 hybrid is a term used in genetics and selective breeding to describe the first filial generation offspring resulting from a cross mating of distinctly different parental types.
Genetic linkage occurs when particular genetic loci or alleles for genes are inherited jointly because the loci are on the same chromosome and thus tend to segregate together during meiosis.
An autosome is a non-sex chromosome.
Sex linkage is the phenotypic expression of an allele that is related to the gender of the individual and is found on the sex chromosomes.
The XY sex-determination system is the sex-determination system found in humans, most other mammals, and some insects.
Heritability is the proportion of phenotypic variation in a population that is attributable to genetic variation among individuals.
Zygosity describes the similarity or dissimilarity of DNA between homologous chromosomes at a specific allelic position or gene.
A phene is an individual characteristic or trait which can be possessed by an organism, such as eye colour or height, or any other observable characteristic.
Complete linkage is defined as the state in which two loci are so close together that alleles of these loci are virtually never separated by crossing over.
A true breeding organism is an organism having certain biological traits which are passed on to all subsequent generations when bred with another organism of the same type for the same traits.
A Barr body is the inactive X chromosome in a female cell.
Penetrance is a term used in genetics describing the proportion of individuals carrying a particular variation of a gene that also express the particular trait.
A norm of reaction describes the pattern of phenotypic expression of a single genotype across a range of environments.
Pleiotropy occurs when a single gene influences multiple phenotypic traits.
A phenome is the set of all phenotypes expressed by a cell, tissue, organ, organism, or species.
Complementation refers to a relationship between two different strains of an organism which both have homozygous recessive mutations that produce the same phenotype.
Alleles have identity by type if they have the same phenotypic effect.
X-inactivation (also called lyonization) is a process by which one of the two copies of the X chromosome present in female mammals is inactivated.
Polygenic inheritance, also known as quantitative or multifactorial inheritance refers to inheritance of a phenotypic characteristic that is attributable to two or more genes and their interaction with the environment.
A haplogroup is a large group of haplotypes, which are series of alleles at specific locations on a chromosome.
A haplotype is a combination of alleles at multiple linked loci that are transmitted together.
The term heterosis, also known as hybrid vigor or outbreeding enhancement, describes the increased strength of different characteristics in hybrids.
Haploinsufficiency occurs when a diploid organism only has a single functional copy of a gene and the single functional copy of the gene does not produce enough of a gene product to bring about a wild-type condition.
A null allele is a mutant copy of a gene that completely lacks that gene's normal function.
Quantitative trait loci are stretches of DNA that are closely linked to the genes that underlie a trait in question.
Canalization is a measure of the ability of a genotype to produce the same phenotype regardless of variability of its environment.
A hypostatic gene is one whose phenotype is masked by the expression of an allele at a separate locus, in an epistasis event.
Malecot's coancestry coefficient refers to an indirect measure of genetic similarity of two individuals.
Ousiotype is a term defined in segregation analysis to describe whether a phenotype is due to common factors, rare or both.
The X0 sex-determination system is a system that grasshoppers, crickets, roaches, and some other insects use to determine the sex of their offspring.
The ZW sex-determination system is a system that birds, some fish, and some insects (including butterflies and moths) use to determine the sex of their offspring.
The haplodiploid sex-determination system determines the sex of the offspring of many Hymenopterans (bees, ants, and wasps), and coleopterans (bark beetles).
 The WikiPremed MCAT Course is a comprehensive course in the undergraduate level general sciences. Undergraduate level physics, chemistry, organic chemistry and biology are presented by this course as a unified whole within a spiraling curriculum. Please read our policies on Privacy and Shipping & Returns. Contact Us. MCAT is a registered trademark of the Association of American Medical Colleges, which does not endorse the WikiPremed Course. WikiPremed offers the customers of our publications or our teaching services no guarantees regarding eventual performance on the MCAT. The WikiPremed MCAT Course is a comprehensive course in the undergraduate level general sciences. Undergraduate level physics, chemistry, organic chemistry and biology are presented by this course as a unified whole within a spiraling curriculum. Please read our policies on Privacy and Shipping & Returns. Contact Us. MCAT is a registered trademark of the Association of American Medical Colleges, which does not endorse the WikiPremed Course. WikiPremed offers the customers of our publications or our teaching services no guarantees regarding eventual performance on the MCAT.

WikiPremed is a trademark of Wisebridge Learning Systems LLC. The work of WikiPremed is published under a Creative Commons Attribution NonCommercial ShareAlike License. There are elements of work here, such as a subset of the images in the archive from WikiPedia, that originated as GNU General Public License works, so take care to follow the unique stipulations of that license in printed reproductions. |